FISH fluorescence in situ hybridization technology and spinning disk confocal microscopy system
1.Introduction
FISH (Fluorescence In Situ Hybridization) is an important method for locating specific nucleic acid sequences in cells or tissues. By labeling DNA or RNA probes and combining with fluorescence microscopy, FISH technology can accurately locate and detect specific genes, chromosome structures and functional regions at the cellular level, and has important application value for research in the fields of cytogenetics, oncology, developmental biology, etc.
2.FISH fluorescence in situ hybridization
The principle of fluorescence in situ hybridization technology is based on the complementary pairing characteristics of nucleic acids. When two single-stranded nucleic acid fragments exist under suitable conditions, they can combine through hydrogen bonds to form stable double-stranded molecules, forming DNA-DNA, DNA-RNA or RNA-RNA double-stranded structures. In fluorescence in situ hybridization, the probe is a labeled DNA or RNA fragment. These labels can be non-radioactive substances such as radioactive isotopes, fluorescein, biotin, digoxin, etc. By combining the fluorescently labeled DNA probe with the specific DNA sequence in the sample to be tested, the position of these specific DNA sequences can be located and detected in the cell. [1] [2]
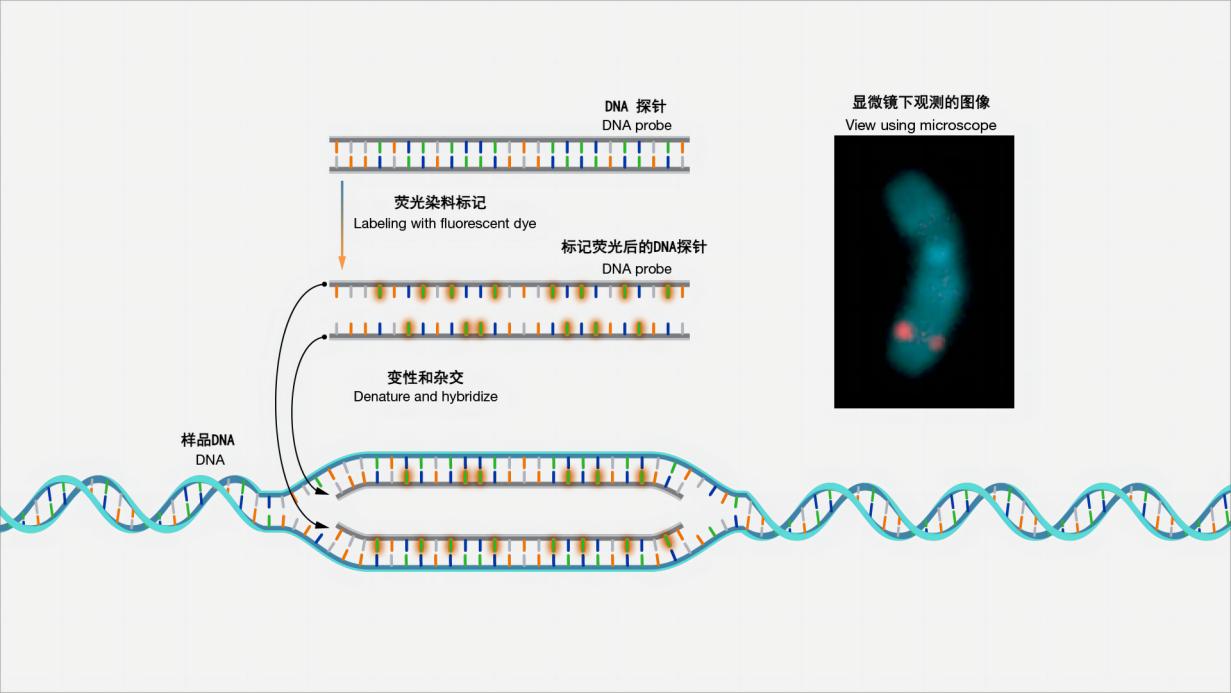
Figure 1 Schematic diagram of fluorescence in situ hybridization
Fluorescence in situ hybridization, or FISH for short, is a laboratory technique used to detect and locate specific DNA sequences on chromosomes. In this technique, the complete chromosome set from an individual is fixed on a glass slide and then exposed to a "probe" - a small piece of purified DNA labeled with a fluorescent dye. The fluorescently labeled probe will find and bind to the matching sequence in the chromosome set. With the help of a special microscope, the chromosomal and subchromosomal locations where the fluorescent probe binds can be seen. (Image and explanation source: Fluorescence In Situ Hybridization (FISH) (genome.gov) )
Fluorescence in situ hybridization was originally developed to detect specific DNA sequences in mammalian chromosomes, and was later applied to the study of plant chromosomes. By using fluorescently labeled DNA probes to bind to specific DNA sequences in the sample to be tested, a fluorescent signal can be formed in the cell, making the specific DNA sequence visible in the cell. The commonly used detection method is fluorescence signal detection under a fluorescence microscope, observing and locating the position of the target nucleic acid sequence under an optical or electron microscope, thereby performing quantitative and positional analysis of gene expression in cells or tissues. This technology helps to study the structure and function of chromosomes, as well as detect chromosomal abnormalities and genetic variations. [1] [3]
Such technology provides researchers with a powerful tool to deeply explore genomic information in cells and tissues. By observing and locating specific DNA sequences, researchers can better understand the mechanisms of gene expression regulation, chromosome structure and function, etc. At the same time, fluorescent in situ hybridization technology also plays an important role in medical diagnosis and genetic research. For example, it can be used to detect chromosomal abnormalities or gene mutations in tumor cells, providing an important basis for the diagnosis and treatment of diseases.
3.FISH technology application cases
FISH is a recognized method for evaluating HER2 gene status in breast cancer. Detection of HER2 status in breast cancer is a prerequisite for screening patients who receive anti-HER2 targeted therapy (trastuzumab and lapatinib, etc.). The use of FISH technology to evaluate HER2 status in breast cancer is considered to be the gold standard for formulating a reasonable systemic treatment strategy for patients with early or advanced breast cancer. Referring to the results of immunohistochemistry, doctors determine the potential amplified invasive tumor tissue cells, and then use fluorescence microscopy to observe and count the number of red HER2 and green CEP18 (chromosome 17 centromere) fluorescent signals in the cell nucleus. According to the 2018 American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) breast cancer HER2 detection guidelines, clinically, it is required to quantitatively count the nuclear signals of at least 20 cancer cells with complete boundaries, and to grade them according to the two indicators of NHER2/NCEP17 ratio (where N is the number) and HER2 average copy number (CHER2). When NHER2/NCEP17 ≥ 2.0 and CHER2/Ncell ≥ 4.0, this condition is considered FISH positive, where Ncell is the number of cells. [6] [8]
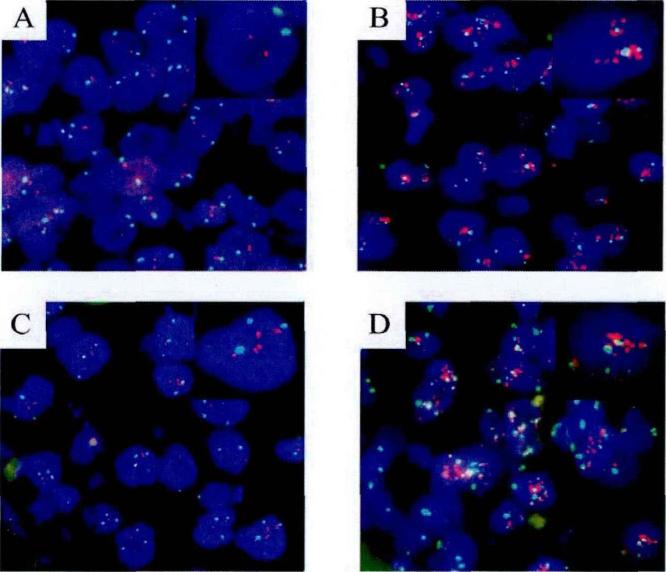
Figure 2 Detection of GER2 gene amplification status using a conventional fluorescence microscope [8]
HER2 gene status was determined by FISH. A, HER2 gene was not amplified, without polysomy 17. B, HER2 gene was amplified, with polysomy 17. C, HER2 gene was not amplified, without polysomy 17. D, HER2 gene was amplified, with polysomy 17. (Image and explanation source: Ma Li. HER2 gene detection, clinical pathological characteristics analysis and epidemiological study of breast cancer in China [D]. Peking Union Medical College, 2012.)
4.Imaging Solutions Comparison
Summarize:
| Observation method |
Advantage |
Shortcoming |
| Widefield fluorescence microscopy |
- One of the main imaging tools used for FISH pathology diagnosis.
- Relatively simple to use and low cost.
- FISH spot coordinates can be easily determined in cells.
|
- Limited by the optical diffraction limit, the imaging resolution is low.
- Fluorescent probes that are closer than the resolution within the nucleus cannot be accurately counted.
- -Not suitable for imaging tissue sections.
|
| Laser scanning confocal microscopy |
- Improved imaging signal-to-noise ratio, better imaging quality compared to wide-field fluorescence microscopy.
- Multiple synaptic structural markers can be visualized simultaneously by fluorescent immunohistochemistry.
|
- The imaging speed is slow and not suitable for practical FISH diagnostic applications.
- The quantum efficiency of using photomultiplier tubes is relatively low.
|
| Spinning disk confocal microscope |
- Has faster imaging speed and higher imaging resolution
- Excellent tool for simultaneously quantifying the amounts of multiple synaptic components and estimating protein levels in tissue sections.
- Faster and better 3D imaging.
- CCD cameras have higher quantum efficiency and higher image quality.
- Reduce photobleaching and phototoxicity.
|
- The spatial resolution is slightly lower than that of point scanning confocal microscopy.
|
Detailed introduction:
Widefield fluorescence microscopy and fluorescence pathology slide scanners are currently the main imaging tools for FISH pathology diagnosis. However, due to the limitation of optical diffraction limit, when using an excitation light with a wavelength of 561 nm, the imaging resolution of FISH probes is about 300 nm. When the distance between two or more fluorescent probes in the cell nucleus is less than the imaging resolution, they cannot be accurately counted. To achieve accurate statistics of HER2 gene levels in positive patients, higher resolution imaging technology is required. In addition, widefield fluorescence microscopy is not suitable for imaging tissue sections because too much out-of-focus blur will be detected. For example, the chromosome region stained by FISH. Although the z-coordinate of the FISH point can be easily determined in a widefield microscope, the chromosome boundary is too blurred and unclear. Widefield microscopy is also unable to accurately determine the 3D cell nuclear boundary based on the counterstained image because there is a lot of out-of-focus haze at the top and bottom of the cell nuclear image. [5] [6]
Laser scanning confocal microscopy (CLSM) uses physical pinholes to filter out out-of-focus signals, significantly improving the imaging signal-to-noise ratio compared with wide-field fluorescence microscopy. Ram et al. used confocal fluorescence microscopy to locate DNA fragments of chromosomal targets and quantitatively calculated spatial distances, and proposed an algorithm for segmenting and detecting 3D FISH spots.[6] The advent of CLSM has significantly improved the ability to simultaneously visualize multiple synaptic structural markers via fluorescent immunohistochemistry. The advantage of evaluating multiple markers simultaneously is that it is possible to determine the frequency with which different antigens colocalize. Additionally, the use of fluorescence allows determination of relative antigen concentrations within identified structures. While CLSM is an excellent research tool for determining the density of labeled synaptic components, it is not necessarily the best tool for quantifying labeled antigen. Since CLSM uses point scanning for imaging, the imaging speed is slow and it is not widely used in actual FISH diagnosis . Although widefield fluorescence microscopy should ideally be used for quantitative fluorescence measurements (Murray et al., 2007; Swedlow et al., 2002), widefield fluorescence microscopy is not suitable for imaging tissue sections because too much ionization would be detected. Blurred. Between CLSM and conventional microscopy, spinning disk confocal microscopy collects 10 times more information than CLSM (Sandison and Webb, 1994).
Spinning disk confocal microscopy is an exceptional tool for quantifying the amounts of multiple synaptic components simultaneously and for estimating protein levels in synaptic structures in tissue sections. In addition, the CCD cameras used for image detection in spinning disk confocal microscopy have high quantum efficiencies (typically over 90%), whereas the photomultiplier tubes used in most CLSMs have relatively low quantum efficiencies (approximately 25%) (Murray et al., 2007; Sandison and Webb, 1994; Shaw, 2006; Wang et al., 2005). The process of collecting 3D image stacks using a spinning disk confocal is much faster than that using a CLSM. The increased speed reduces the amount of photobleaching that occurs above and below the focal plane, an effect that is magnified when imaging thick tissue sections because a large number of focal planes are being imaged . In addition, the increased speed facilitates the collection of large data sets required for most stereological sampling schemes. [7]
5.Imaging solutions and imaging results from YK
Based on the vision of increasing laboratory productivity without compromising data quality, our company developed the SpinDisk series, which provides the most affordable solution for fast and gentle confocal imaging.

Figure 3: SpinDisk Basic
Fluorescence microscopes, the SpinDisk Basic has higher imaging speed and resolution, enabling more accurate three-dimensional imaging while reducing the effects of light quenching and fluorescence signal attenuation on image quality.
The challenges that FISH (fluorescence in situ hybridization) technology may face during observation mainly include:
- Weak fluorescence signal: Fluorescence signal may be affected by many factors, such as the affinity of the probe, the way the sample is processed, etc., resulting in weak signal and difficult to observe and analyze.
- Background noise interference: Background noise may exist during sample preparation, such as nonspecific binding or spontaneous luminescence, which mixes with the target signal and interferes with the clarity and accuracy of the image.
- Multi-channel imaging overlay: When using multiple fluorescence channels, signals from different channels may overlap with each other, making it difficult to analyze and identify specific signals.
-
SpinDisk Basic confocal microscope system from CYC offers a range of solutions:
- High-brightness laser light source: The system uses a multi-channel continuous multi-mode laser (four-in-one laser) as a light source to provide high-brightness laser to enhance the intensity of the fluorescence signal, making the signal easier to observe and analyze. The advantage of the four-in-one laser system is that it can provide a multi-wavelength laser light source, which can simultaneously excite multiple fluorescent dyes or markers to obtain more comprehensive information in one experiment. This is of great significance for multi-color fluorescence imaging, co-localization and co-expression research of cells and tissues.
- One-key switching of multi-channel excitation: The system is equipped with an electric filter wheel. The electric filter wheel and multi-channel laser are controlled by software to achieve a one-key switching function for multi-channel excitation, which can quickly adjust the excitation and emission channels to avoid signal superposition problems in different channels.
- High-quality filters and background suppression technology: The system is equipped with high-quality filters that can effectively filter out background noise, and uses turntable supporting software developed by MK , background suppression technology such as intelligent background correction algorithm, etc. to improve image clarity and signal-to-noise ratio.
- High-sensitivity detector: The system uses a high-sensitivity detector, BSI sCOM camera , which can effectively capture fluorescence signals and provide high-quality imaging results.
Combining SpinDisk Basic with FISH technology, these solutions can overcome some of the limitations of traditional imaging solutions and achieve higher quality FISH imaging with rich features , including but not limited to :
- Large image stitching : The system has a large image stitching function that can stitch multiple local images into one large image, thereby expanding the imaging range and obtaining a larger field of view.
- Real-time preview and image storage : The system provides real-time image preview function, allowing users to observe the fluorescence signal of the sample in real time and collect images. At the same time, users can directly store the observed images for subsequent analysis and processing.
- Multi-channel imaging and fusion : The system supports multi-channel fluorescence imaging, which can capture images of multiple fluorescence channels at the same time. This allows users to detect multiple target DNA sequences or markers on the same sample at the same time, thereby obtaining more comprehensive information. In addition, the system also provides image fusion function, which superimposes images of different channels for easy observation and analysis.
- Image processing : The system is equipped with image processing software that can adjust, enhance and correct images to improve image quality and contrast, helping users observe sample structures and signals more clearly.
- 3D reconstruction : The system supports 3D reconstruction of imaged samples, that is, reconstructing the 3D sample structure from a series of images. This allows users to observe the distribution and location of target DNA sequences throughout the sample volume, thereby gaining a more comprehensive understanding of the spatial structure of the sample.
- Multicolor fluorescence localization processing : The system can perform localization processing for multiple different fluorescent markers, that is, determine the location and distribution of different fluorescent signals. This is very important for FISH experiments that simultaneously detect multiple targets or markers.
- Z-stack data processing : The system supports Z-stack data processing, which is to acquire a series of images along the Z axis and then stack them to reconstruct the three-dimensional structure of the sample. This helps users to have a more comprehensive understanding of the three-dimensional morphology and spatial distribution of the sample.
- Image analysis : The system provides image analysis tools that can perform quantitative analysis and measurement on images, such as calculating the intensity, distribution and correlation of fluorescence signals, thereby helping users to quantitatively analyze experimental results.
- Imaging data management : The system has imaging data management function, which can store, manage and retrieve the collected images and data, ensure the security and integrity of the data, and facilitate the user's subsequent data processing and review.
- 3D imaging rendering : The system supports rendering and display of three-dimensionally reconstructed samples, and can achieve a variety of different three-dimensional imaging effects, so that users can more intuitively observe and understand the three-dimensional structure of the sample.
- Hardware control : The system has complete hardware control functions, which can accurately control hardware parameters such as light source, filter, lens, etc. to ensure the stability and accuracy of imaging.
- Multi-dimensional automatic scanning : The system supports multi-dimensional automatic scanning, which can automatically scan samples in multiple dimensions according to the scanning parameters set by the user, thereby achieving more comprehensive imaging coverage and more efficient data acquisition.
SpinDisk Basic imaging demonstration:


Figure 4 405/488/525 three channels 60X 1.2NA cervical cancer cell FISH probe sample acquisition
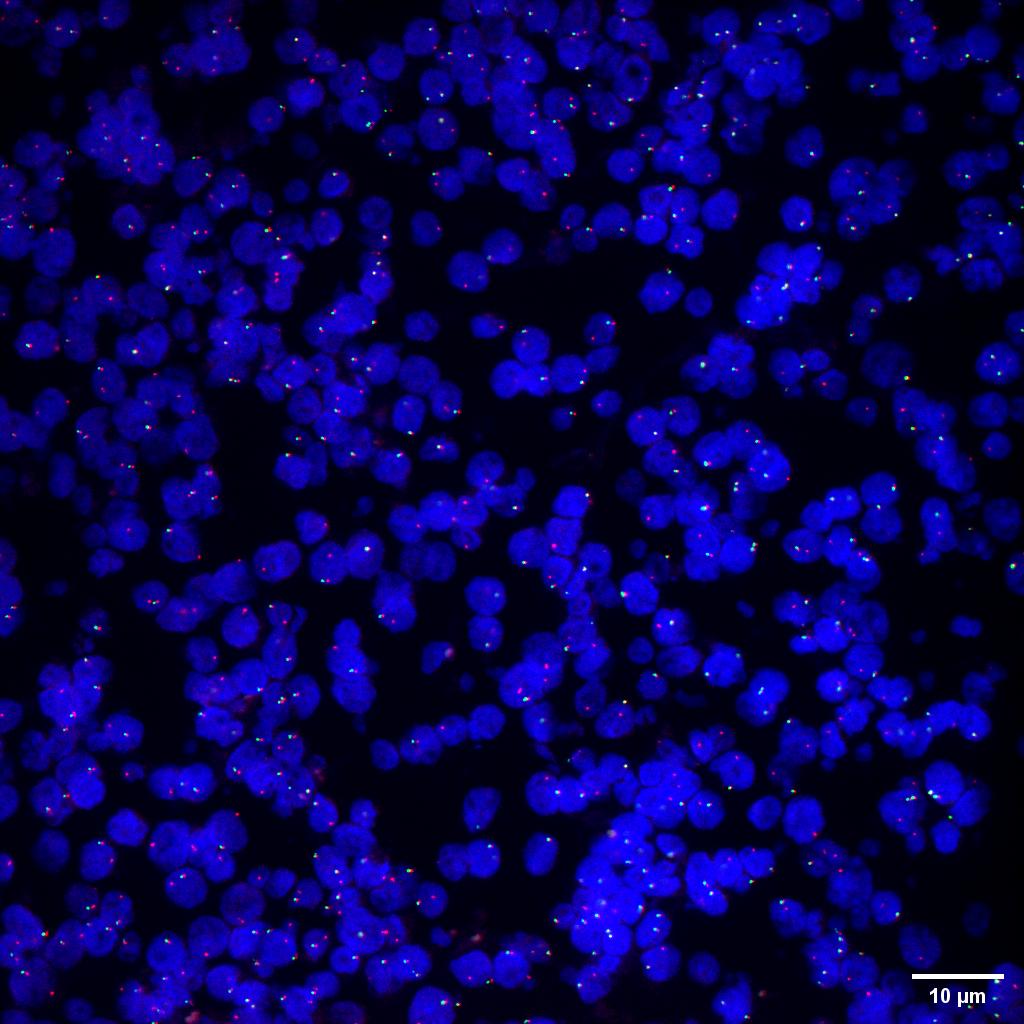
Figure 5 SpinDisk Basic-60X 1.2NA-405/488/525-cervical cancer cell FISH probe sample multicolor fusion
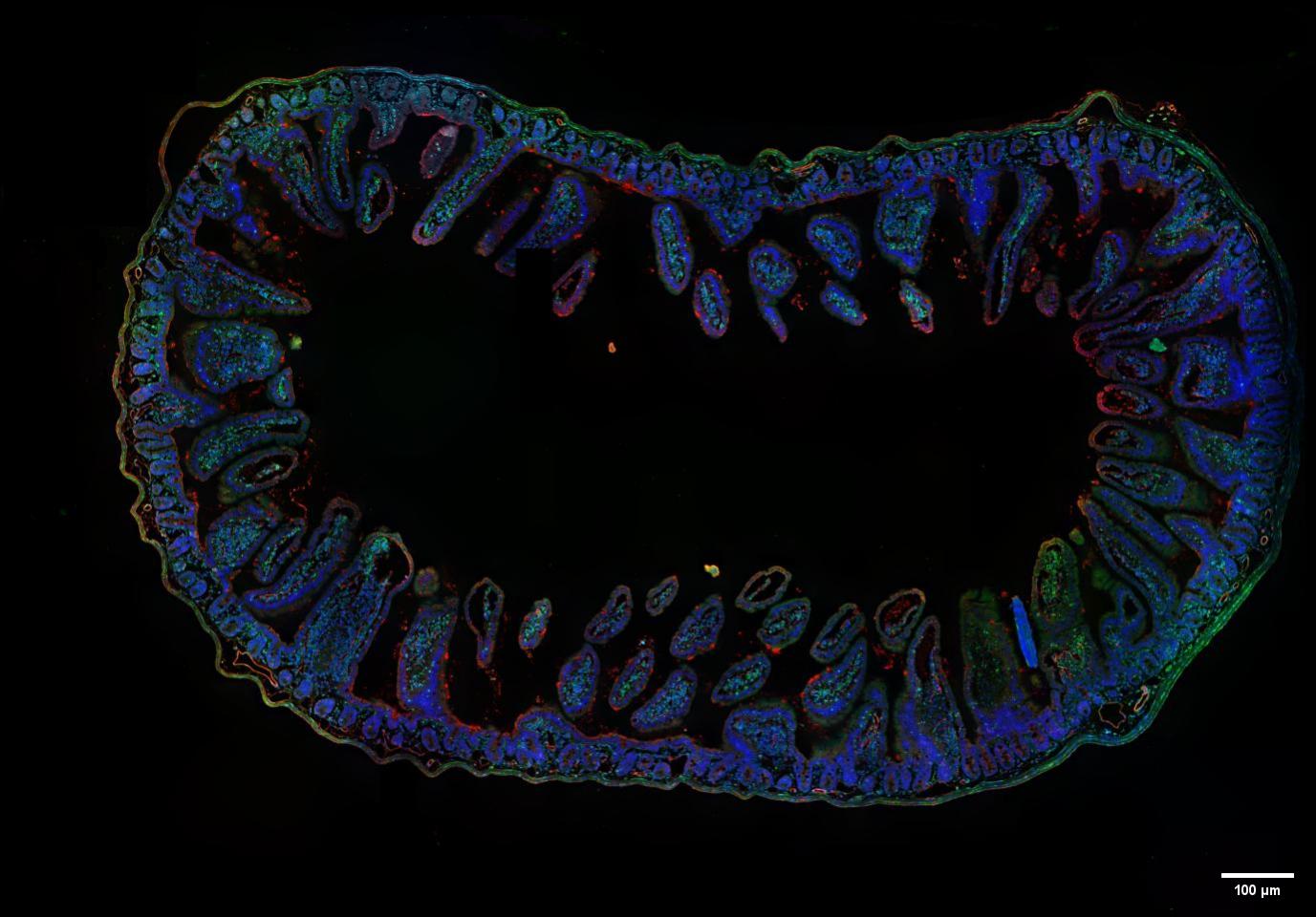
Figure 6 SpinDisk Basic-20X 0.5NA-405/488/525-Multi-color fusion and large image stitching of mouse intestinal tissue embedded sections- the full image size 1.8mmX1.25mm is stitched from 56 partial images
The SpinDisk Basic confocal system combines a 20X 0.5NA objective lens and multi-channel lasers (405nm, 488nm and 525nm), providing multi-color fusion and large image stitching functions, providing users with a more comprehensive and clearer imaging solution.
Using the SpinDisk Basic's spinning disk confocal mode, we can quickly capture fluorescence images of different channels to obtain the fluorescence signals of the target sample at different wavelengths. Through optimized image processing algorithms, these multi-channel images can be accurately fused together to form a colorful composite image, showing the multi-color fluorescent labeling and distribution of the sample.
In addition, SpinDisk Basic also has a powerful large image stitching function, which can stitch multiple local images into one large image, expanding the imaging range and improving the imaging resolution and field of view. This provides users with more comprehensive and detailed sample information, helping them to better understand and analyze the structure and characteristics of the sample. In the process of stitching images, we use an optimized algorithm to ensure the continuity and accuracy of the image.
Company Profile:
Founded in Singapore in 2019, MC Instruments has an excellent R&D team. The core members have decades of experience in optical technology and industry. They focus on the innovation and application of microscopy technology and have successively developed high-end microscopy systems with Nanyang Technological University (NTU), National University of Singapore (NUS) and A-Star. We focus on confocal, super-resolution microscopy and other related fields, and are committed to producing high-end microscope products for scientific research and industrial grade. Our core product is the high-resolution microscope series, which uses excellent optical technology and image processing algorithms, and has excellent imaging performance to meet customers' needs in high-resolution and fast imaging.
6.Citations
[1]Shakoori AR. Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations. 2017 Feb 10:343–67. doi: 10.1007/978-81-322-3673-3_16. PMCID: PMC7122835.
[2]Jin, L. and Lloyd, RV (1997), In situ hybridization: Methods and applications. J. Clin. Lab. Anal., 11: 2-9. https://doi.org/10.1002/(SICI)1098- 2825(1997)11:1<2::AID-JCLA2>3.0.CO;2-F
[3]Liu Yuling, Liu Linjie, Peng Renhai. Development of fluorescence in situ hybridization technology and its application in plant genome research[J]. Molecular Plant Breeding, 2018, 16(17): 5696-5703. DOI: 10.13271/j.mpb. 016.005696.
[4]Fluorescence In Situ Hybridization (FISH)By: Clare O'Connor, Ph.D. (Biology Department, Boston College) © 2008 Nature Education Citation: O'Connor, C. (2008) Fluorescence in situ hybridization (FISH). Nature Education 1(1):171
[5]Kozubek, M., Kozubek, S., Lukášová, E., Bártová, E., Skalníková, M., Matula, P., Matula, P., Jirsová, P., Cafourková, A. and Koutná, I. ( 2001), Combined confocal and wide-field high-resolution cytometry of fluorescent in situ hybridization-stained cells. Cytometry, 45: 1-12.
[6]Wu Yin, Liang Yong, Zhang Jie, et al. Structured light illumination super-resolution imaging and counting of fluorescence in situ hybridization amplified probes[J]. Laser & Optoelectronics Progress, 2024, 61(4):0411009.DOI: 10.3788/LOP231182.
[7]Kenneth N. Fish, Robert A. Sweet, Anthony J. Deo, David A. Lewis,An automated segmentation methodology for quantifying immunoreactive puncta number and fluorescence intensity in tissue sections, Bran Research,Volume 1240,2008,Pages 62-72,ISSN 0006-8993
[8]Ma Li. HER2 gene detection, clinical pathological characteristics analysis and epidemiological study of breast cancer in China[D]. Peking Union Medical College, 2012.